Exploring Complex Phenotypes with Genomics: Dalmatian Deafness and Labrador Retriever Anxiety
Introduction
Our focus today will consider two complex traits that are undergoing investigation in our laboratory. Our goal is to discover critical genetic drivers of these traits that might be used to improve dog welfare either through better physical management or through tools to enhance breeding strategies. Our laboratory focuses on the use of within-breed gene mapping as a strategy to uncover genes that impact common phenotypes. This presentation will focus on inherited deafness and a common behaviour disorder. Both traits are expected to be impacted by multiple genes as well as environment and other non-genetic factors (such as random chance).
Dalmatian Deafness
One of the most common forms of peripheral hearing loss in dogs is congenital sensorineural deafness (CSD). Dalmatians are frequently represented in lists of breeds that exhibit CSD, and prevalence in the breed is reported to be 16–29.9%. CSD is known to be inherited; and heritabilities have been reported that suggest selection will effectively improve it. CSD in the Dalmatian breed is classified as non-syndromic and can occur in both unilateral and bilateral forms.
Dalmatian deafness has long been presumed to be related to the extreme white coat coloration in the breed. While Dalmatians are somewhat less “white” than other breeds with the extreme white coat colour (such as the bull terrier), they exhibit a higher rate of deafness in pups. This suggests that the coat pigmentation alone is not responsible for the phenotype.
Methods
Deafness was assessed through the application of brainstem auditory evoked response (BAER) testing of pups between six and eight weeks of age. Several different BAER testing devices can be employed for this purpose and not are all equally reliable. For our study, a veterinary neurology specialist (Child) used a Synergy on Nicolet EDX System (Natus Medical Incorporated, Pleasanton, California, USA) with a repetitive click stimulus that was applied to the pup via inserted earphones. The device was used to ascertain normal hearing, unilateral deafness or bilateral deafness.
Either buccal swab or blood samples were collected from the pups with the permission of the owners and DNA was extracted. One hundred seventy-two Dalmatian DNA samples were available for analysis and were genotyped using Illumina Canine Genotyping arrays (Neogen Inc., Nebraska, USA). Genome-wide association mapping applied a quantitative model using the Plink software (Purcell et al. 2007).
Results
Our results validated the findings from an earlier study in the Dalmatian breed that showed complex association including association with the white-markings locus including the gene microphthalmia-associated transcription factor (MITF).
Conclusion
The precise mutations that are impacting the phenotype are yet to be identified but a risk marker haplotype has been established that concords with the earlier finding.
Figure 1
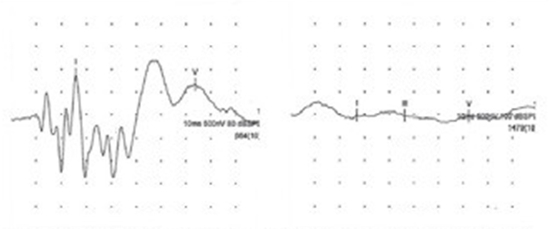
a. Nicolet EDX BAER trace taken from an ear with normal hearing. Note the pattern and amplitude of peaks and troughs. b. BAER trace taken from a completely deaf ear demonstrating an absence of distinct waveform (images reproduced with permission from owner).
Canine Separation-Related Distress
A common problem in companion dogs is separation related distress (also called separation anxiety). As affected dogs are often vocal or destructive when left alone, the disorder results in the euthanasia or relinquishment of many dogs world-wide.
To map genes underlying the disorder, we have concentrated on the inheritance of this condition in Labrador retrievers. The disorder manifests in dogs of many different breeds as well as cross-breeds—perhaps as a result of humans selecting for dogs that are very attached to us. We sought to use a within-breed mapping strategy, and this was facilitated by choosing a breed that is readily accessible as one of the ten most popular dog breeds in Australia (and indeed in many countries world-wide).
Methods
Separation-related distress was assessed by an owner-based questionnaire based on C-BARQ. Dogs had lived with their owners for at least 6 months. Owners reported on the frequency of 8 behaviours commonly seen in separation-related distress, and the 8 scores averaged to give an overall score for each dog. We validated the questionnaire by comparing owner responses to an analysis of video footage of dogs left alone.
Buccal swabs were collected from privately-owned dogs by ourselves or the owners. DNA was extracted and genotyped using Illumina Canine Genotyping arrays (Neogen Inc., Nebraska, USA). A quantitative model of genome-wide association analysis was conducted using Plink software (Purcell et al. 2007). We had 90 Labrador retrievers with both questionnaire and genotyping data that were analysed.
Results
Of the top ten most significant markers, three were within an 80 kilobase region on chromosome 1 and four within a 2 megabase region on chromosome 12. Genes involved in the HPA-stress axis, schizophrenia and social behaviour are located within and adjoining these areas of interest. We have not yet identified candidate genes or mutations.
Figure 2
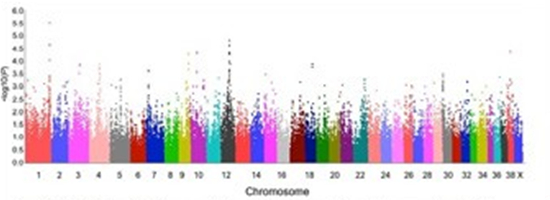
Manhattan plot of genome-wide quantitative association using separation-related behavior scores of 90 Labrador retrievers.
Conclusion
Identifying genes associated with behavioural disorders is challenging. While our work has produced some exciting results, further work is needed to validate our findings and identify the genes involved in the development of separation-related distress.